The liver #transcriptome is highly sensitive to metabolic cues. By characterizing nuclear protein #OGlcNAcylation in mouse #liver during the day, this study shows how shifts in a nutrient-sensitive #PTM may shape transcriptional regulation @PLOSBiology https://plos.io/3KuS7m1
#transcriptome
Wang et al. explore #adaptation to #karst geology in the medicinal plant Hemiboea subcapitata using a #chromosome-level #genome and #transcriptome analysis, revealing effects on flavonoid metabolism. #plantscience #evolution #botany
@wileyecolevol @wileyplantsci
https://doi.org/10.1111/jse.70007
Pipeline release! nf-core/denovotranscript v1.2.1 - nf-core/denovotranscript v1.2.1!
Please see the changelog: https://github.com/nf-core/denovotranscript/releases/tag/1.2.1
#denovoassembly #rnaseq #transcriptome #nfcore #openscience #nextflow #bioinformatics
The #spider #Tibellus #oblongus (#Philodromidae) has a #Holarcticdistribution and is common in Central Europe, thus not being protected. It prefers #sunexposed #habitats, whether dry or moist, and is a free #hunter that is very well #camouflaged due to its elongated #bodyshape. Y. Korolkova et al. (2023) examined the #venomgland #transcriptome and a venom proteome analysis.
© #StefanFWirth #Rosenthal #Berlin 2025
Ref
Y. Korolkova et al. (2023)
https://doi.org/10.1038/s41597-023-02703-0
Photos
© S.F. Wirth
Differential gene expression during early development in recently evolved and sympatric Arctic charr morphs
old paper on the development of sympatric morphs
#arcticcharr #transcriptome #rnaseq #fish #evolution #sympatry
#peerj #biology
https://peerj.com/articles/4345/
#Microbiota transplantation offers new hope against #cotton #leaf_curl #disease.
#CLCuD #rhizosphere #phylosphere #Gossypium # #microbiome #transcriptome
https://phys.org/news/2025-03-microbiota-transplantation-cotton-leaf-disease.html
Are you ready for #EMBLEpitrans? Prepare to deep dive into advances in RNA modification functions, their key roles in gene expression, disease, and transformative therapies. Be part of the community🔬💡
📩 Submit your abstract by 5 August
🗓️ 28 – 30 October
📍 EMBL Heidelberg and Virtual
➡️ https://s.embl.org/etc25-01-ma
#EMBLEvents #geneexpression #RNAmodification #mRNA #RNA #transcriptome
Pipeline release! nf-core/denovotranscript v1.2.0 - nf-core/denovotranscript v1.2.0!
Please see the changelog: https://github.com/nf-core/denovotranscript/releases/tag/1.2.0
#denovoassembly #rnaseq #transcriptome #nfcore #openscience #nextflow #bioinformatics
Random question for #transcriptome #bioinformatics folks - is there ever a reason to screen transcriptome assemblies (lots of small contigs) against database using ANI tools (i.e. skani)?
Curious to know if there's a usage case for it.
Do or die: How #soybeans tackle #nematode invaders.
#agriculture #transcriptome #RNA
https://phys.org/news/2025-01-die-soybeans-tackle-nematode-invaders.html
I've been seeing a few #transcriptome wide association studies (TWAS) papers lately. This is another example, from #sorghum, combining GWAS and TWAS to study flowering time. Bonus - I imagine that there's a lot more value that can be gained from the 822 #RNAseq samples made publicly available
https://www.biorxiv.org/content/10.1101/2024.12.12.628249v1.full.pdf
Pipeline release! nf-core/denovotranscript v1.1.0 - nf-core/denovotranscript v1.1.0!
Please see the changelog: https://github.com/nf-core/denovotranscript/releases/tag/1.1.0
#denovoassembly #rnaseq #transcriptome #nfcore #openscience #nextflow #bioinformatics
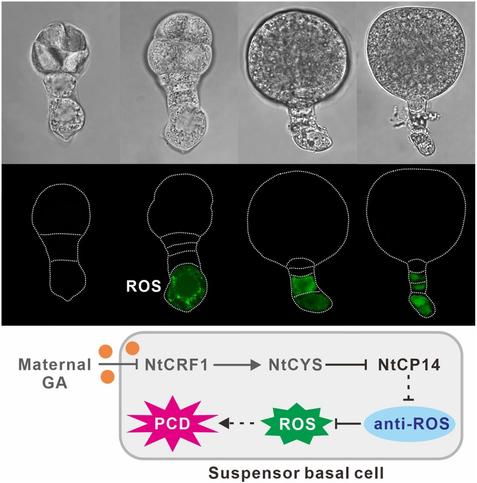
Luo et al. use comparative #transcriptome analysis with isolated two-celled #proembryos to explore the potential regulatory network underlying NtCP14-triggered programed #cell death in #embryos.
https://doi.org/10.1111/jipb.13795
@wileyplantsci
#PlantSci #JIPB #plant #development #embryogenesis #botany
Human molecular map contributes to understanding of disease mechanisms
https://www.sciencedaily.com/releases/2024/09/240911175947.htm
Roadmap to molecular human linking multiomics w. population traits, diabetes subtype
https://www.nature.com/articles/s41467-024-51134-x
6,304 quantitative molec. traits
1,221,345 genetic variants
methylation @ 470,837 DNA CpG sites
gene expression: 57,000 transcripts
#metabolome #connectome #interactome #genotype #phenotype #IndividualVariation #transcriptome #multiomics #omics #disease #MolecularEpidemiology #GeneExpression
Pipeline release! nf-core/denovotranscript v1.0.0 - nf-core/denovotranscript v1.0.0!
Please see the changelog: https://github.com/nf-core/denovotranscript/releases/tag/1.0.0
#denovo-assembly #rna-seq #transcriptome #nfcore #openscience #nextflow #bioinformatics
#Genetic signatures of #domestication identified in #pigs and #chickens.
#agriculture #anthropology #transcriptome
https://phys.org/news/2024-08-genetic-signatures-domestication-pigs-chickens.html
Transcriptome- and proteome-wide effects of a circular RNA encompassing four early exons of the spinal muscular atrophy genes. https://doi.org/10.1038/s41598-024-60593-7 #SpinalMuscularAtrophy,Sma #SurvivalMotorNeuron,Smn #CircularRna,Circrna #Proteome #Transcriptome
You know what really 🪲#bugs🪲me? Not having a stable framework for understanding the #evolutionary history of #Cucujiformia...
...Oh, hello there, Li et al.!
https://doi.org/10.1111/jse.13079
@WileyEcolEvol
#transcriptome #phylogenetics #data #Coleoptera #beetle #insect #entomology #evolution
The #genetic interplay in Impatiens #downy_mildew: A #transcriptome-based approach to enhancing #disease resistance.
https://phys.org/news/2024-05-genetic-interplay-impatiens-downy-mildew.html
Team reports on relationship between contents of #diosgenin and #brassinosteroids in #Dioscorea zingiberensis.
#hypomethylation #transcriptome
https://phys.org/news/2024-04-team-relationship-contents-diosgenin-brassinosteroids.html