Moez Dawood, an MD/PhD student at Baylor College of Medicine, created the AllofUs 250k annotator for OpenCRAVAT to improve genomic research by integrating diverse data, including ancestry-specific allele frequencies. You can read more about Moez's experience developing the annotator here: https://www.opencravat.org/bringing-genetic-diversity-to-the-forefront-moez-dawood-on-the-allofus-250k-annotator-for-opencravat/
#annotator #opencravat #bioinformatics #opensource #genomics
#OpenCRAVAT
Would you be interested in organizing or speaking at meetups, writing blog posts, or promoting the project?
Are you part of any communities or organizations where you could share our project? #annotator #opencravat #bioinformatics #opensource #genomics
Can you help test new features or provide feedback on pre-release versions?
Would you be interested in setting up automated test cases or improving test coverage?
Would you be open to helping improve documentation or writing user guides?
Do you have skills in UI/UX design, and would you be interested in helping us improve the user interface?
#annotator #opencravat #bioinformatics #opensource #genomics
We are seeking community members interested in taking part in OpenCRAVAT's future.
Let us know if you might contribute in any of these areas:
Would you be interested in contributing code, bug fixes, or performance improvements?
Are there features or issues you’d like to help implement or debug?
Would you be willing to review pull requests or help triage GitHub issues?🖥️ 🧬 🧪 #annotator #opencravat #bioinformatics #opensource #genomics
🧬 Want to explore your own genome?
Join our free webinar on how to use OpenCRAVAT to annotate & interpret your personal variants.
📅 July 10 | 🕐 1 PM EDT
🎙️ With Jasmine Baker, PhD & Rachel Karchin, PhD
🔗 https://tinyurl.com/3ur6vv35
#Genomics #Bioinformatics #OpenSource #OpenCRAVAT
🧬 Curious about your genome?
Join our free webinar on OpenCRAVAT to explore and annotate your own data!
📅 July 10 – 1 PM EDT
🎙️ Baker & Karchin (JHU)
🔗 https://tinyurl.com/3ur6vv35
#Genomics #Bioinformatics #OpenScience
#annotator #opencravat #bioinformatics #opensource #webinar #genomes 🖥🧪
Discover the power of OpenCRAVAT's cutting-edge annotation features that ensure seamless collaboration among users. Its intuitive design makes it easy for everyone to navigate, while enhancing productivity across the board. Whether you're a beginner or a seasoned pro, OpenCRAVAT opens up a world of possibilities, making your workflow smoother and more efficient.#annotator #opencravat #bioinformatics #opensource #genomics
Discover the power of the All of Us annotator with Open Cravat! This tool leverages the vast allele frequency data from the All of Us Research Program, offering insights into genetic variations across diverse populations. By using this annotator, researchers can delve into non-European genetic diversity and advance precision medicine. Let's drive innovation in healthcare together! #PrecisionMedicine #Genomics #AllOfUs #OpenCravat#annotator #bioinformatics #opensource
Do you have research goals for 2025!? We'd love to hear them! #annotation #OpenCRAVAT #2025 🧪🖥
🧬 New Blog by Dr. Rachel Karchin!
Dr. Karchin shares how OpenCRAVAT and CIViC are addressing transparency and interpretability in variant effect predictors. Discover how calibrated predictors are transforming clinical variant classification.
🔗 Read the full post: https://www.opencravat.org/variant-effect-predictors-in-the-clinic-working-with-calibrated-predictors-in-opencravat/
What are your thoughts on the new ACMG/AMP framework integration? Let us know below!
#Genomics #VariantAnnotation #OpenCRAVAT 🖥️
🌐 On the 10th day of #OpenCRAVAT, my workflow gave to me…
✨ Powerful API Capabilities!
Integrate OpenCRAVAT into pipelines: submit jobs, check statuses, and fetch custom reports programmatically.
Explore the API docs: https://docs.opencravat.org/en/latest/API.html
#12DaysofOpenCRAVAT #Genomics
☁️ Day 9 of #OpenCRAVAT: Run on the Cloud!
No hardware limits, no delays—just efficient genomic analysis anywhere. Learn how to set up OpenCRAVAT on the cloud today.
🔗Cloud Usage: https://docs.opencravat.org/en/latest/Cloud.html
#12DaysofOpenCRAVAT #CloudComputing 🖥️ 🧬 #Genomics #Variants
🔎 Day 8 of #OpenCRAVAT: Eight Data Filtering Options!
Refine your results with precision: filter by gene, variant type, pathogenicity score, or population frequency.
Which filter is most useful for your research? Comment below!
🔗Filtering Tutorial: https://docs.opencravat.org/en/latest/Filter-Tutorial.html
#12DaysofOpenCRAVAT #VariantAnalysis 🧬 🖥️
🌟 On the 6th day of #OpenCRAVAT, my database gave to me…Six Gene-Level Insights!
Which insight is most valuable for your research? CHASMplus, REVEL, PolyPhen-2, gnomAD, ClinVar, or COSMIC?
Explore OpenCRAVAT at opencravat.org
On the 5th day of #OpenCRAVAT, my platform gave to me…
✨ Five Tips for New Users!
New to OpenCRAVAT? Check out the links in this thread. Got questions? Comment below!
🔗 Creating an OC account (web) https://docs.opencravat.org/en/latest/getting_started_web.html#creating-an-opencravat-account-web
#12DaysofOpenCRAVAT #Genomics #OpenData #Variants #Annotation
❄️ On the 4th day of #OpenCRAVAT, my workflow gave to me…
Four Annotation Sources!
Discover how gnomAD4, dbSNP, ClinVar, and COSMIC make your variant analysis powerful. Which do you rely on most?
Link to full list of annotators: https://buff.ly/3OF6sLD
✨ On the 3rd day of #OpenCRAVAT, my tools gave to me…
🎄 Three Custom Report Options!
Choose from TXT, Excel, or Sqlite outputs to fit your workflow. Which report type is your favorite? Reply below!
🔗https://docs.opencravat.org/en/latest/3.-Viewing-Results.html#downloadable-reports
🔗https://docs.opencravat.org/en/latest/3.-Viewing-Results.html#viewing-results
#12DaysofOpenCRAVAT #Genomics #VariantAnalysis
🎁 On the 2nd day of #OpenCRAVAT, my software gave to me…
✨ Two New Example Inputs!
Test OpenCRAVAT with hgvs and dbsnp inputs. Perfect for trying the software before running your data!
🔗https://buff.ly/3BapIgP & https://buff.ly/49l5yNJ
#12DaysofOpenCRAVAT #VariantAnnotation
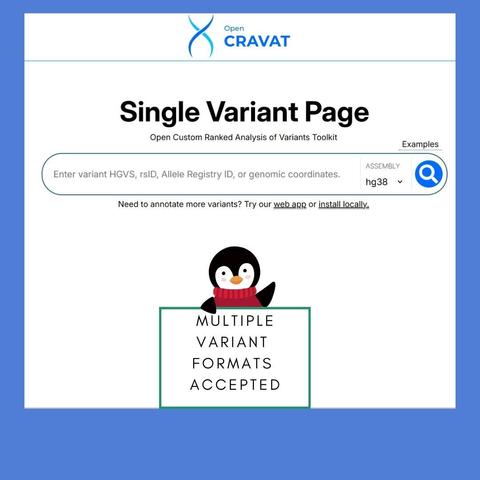
🎄 On the 1st day of #OpenCRAVAT, my analysis gave to me…
✨ A Single Variant Tool for Free!
Quickly annotate any genomic variant right from the new OpenCRAVAT website. Try it now and simplify your workflows!
https://www.opencravat.org/ https://run.opencravat.org/webapps/variantreport/index.html
🌐 Exciting news! OpenCRAVAT has a brand-new website AND logo 🎉. Check it out at https://opencravat.org.
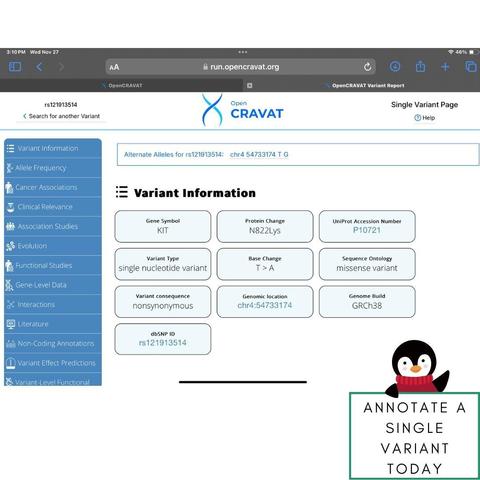
🧬 Explore our Single Variant Tool directly on the landing page—annotate a single variant with ease and see how OpenCRAVAT simplifies genomic analysis! 🚀
Let us know your thoughts! #Genomics #VariantAnalysis #OpenCRAVAT